High-quality cellular imaging is the foundation of accurate biological research, yet noise and artifacts frequently compromise dataset integrity and experimental outcomes.
🔬 Understanding the Challenge: Why Cellular Images Get Messy
Cellular image datasets represent one of the most valuable resources in modern biological research, drug discovery, and medical diagnostics. However, these datasets are notoriously susceptible to various forms of noise and artifacts that can significantly impact downstream analysis. From fluorescence microscopy to phase contrast imaging, researchers constantly battle technical imperfections that obscure the biological signal they’re trying to capture.
The problem isn’t trivial. A single noisy image can skew automated cell counting algorithms, mislead machine learning models, or cause researchers to draw incorrect conclusions about cellular behavior. As high-content screening and automated image analysis become standard practice, addressing image quality issues has transformed from a minor inconvenience to a critical research imperative.
Understanding the sources of these imperfections is the first step toward eliminating them. Noise in cellular imaging can originate from camera sensors, environmental factors, sample preparation issues, or even the fundamental physics of light interaction with biological specimens. Artifacts, on the other hand, often result from optical aberrations, imaging system limitations, or processing errors that introduce false structures into images.
📸 Common Types of Noise in Cellular Imaging
Not all noise is created equal. Different imaging modalities and experimental conditions produce characteristic noise patterns that require specific mitigation strategies.
Shot Noise and Photon Statistics
Shot noise represents the fundamental quantum nature of light detection. When photon counts are low—common in live-cell fluorescence imaging to minimize phototoxicity—the random arrival of photons at the detector creates grainy images. This Poisson-distributed noise is especially problematic in time-lapse imaging where maintaining cell viability requires reducing light exposure.
The signal-to-noise ratio in these conditions depends on the square root of the signal intensity, meaning dim cellular structures suffer disproportionately. Researchers working with weakly expressing fluorescent proteins or studying subcellular compartments with low probe concentrations must develop specialized approaches to extract meaningful information from inherently noisy data.
Read Noise and Dark Current
Camera electronics contribute their own noise signatures. Read noise occurs during the conversion of accumulated charge to digital values, while dark current represents thermal electrons generated even without light exposure. Modern scientific cameras like sCMOS sensors have dramatically reduced these noise sources, but they remain relevant, particularly in long-exposure imaging or when working with older equipment.
Temperature control becomes crucial here. Cooling cameras to -20°C or lower substantially reduces dark current, but introduces operational complexity and cost considerations that not all laboratories can accommodate.
Background Fluorescence and Autofluorescence
Biological samples are rarely perfectly clean optical systems. Cell culture media, mounting solutions, and the cells themselves emit background fluorescence that competes with the specific signal from labeled structures. Autofluorescence from cellular components like NADH, flavins, and lipofuscin can be particularly troublesome in certain wavelength ranges.
This biological noise varies with cell type, culture conditions, and cellular metabolic state, making standardized correction approaches challenging. Red-shifted fluorophores often provide better signal-to-background ratios, but wavelength selection involves tradeoffs with photobleaching rates and spectral overlap in multi-color imaging.
🎯 Recognizing Artifacts That Corrupt Your Data
While noise adds random variations, artifacts introduce systematic distortions that can be even more insidious because they may appear consistent across images, potentially being mistaken for real biological structures.
Optical Aberrations and Field Curvature
Even high-quality microscope objectives suffer from residual aberrations. Spherical aberration causes point sources to appear as extended structures with characteristic halos. Chromatic aberration shifts different wavelengths to different focal planes, complicating multi-color colocalization studies. Field curvature means that flat specimens appear sharp in the center but progressively blur toward image edges.
These effects intensify when imaging through thick specimens or using objectives at wavelengths outside their design specifications. Researchers purchasing objectives should carefully consider correction specifications and ensure compatibility with their specific application requirements.
Uneven Illumination and Vignetting
Perfectly uniform illumination across the entire field of view is surprisingly difficult to achieve. Light source intensity variations, dust on optical elements, and vignetting by apertures create systematic brightness gradients. In widefield fluorescence microscopy, these effects combine with objective collection efficiency variations to produce images that are brighter in the center than at the edges.
This shading artifact compromises quantitative intensity measurements and can confound segmentation algorithms that rely on threshold-based approaches. Without correction, comparing cellular fluorescence intensities between image regions becomes problematic.
Out-of-Focus Light and Haze
Widefield microscopy collects light from throughout the specimen thickness, not just the focal plane. This out-of-focus light appears as haze that reduces contrast and obscures fine details. The problem scales with specimen thickness—imaging cells in 3D culture or tissue sections suffers more severely than monolayer cultures on coverslips.
Confocal and other optical sectioning techniques physically reject out-of-focus light, but introduce their own artifacts and operational constraints. Computational approaches offer alternatives that preserve the gentler imaging conditions of widefield microscopy while recovering optical sectioning capability.
⚙️ Pre-Acquisition Strategies: Prevention Over Cure
The most effective approach to handling noise and artifacts is preventing them from entering your dataset in the first place. Thoughtful experimental design and careful microscope operation pay enormous dividends.
Optimizing Imaging Parameters
Camera exposure time, excitation intensity, and detector gain represent interconnected parameters that profoundly influence image quality. Longer exposures collect more photons, improving shot-noise-limited signal-to-noise ratios, but increase photobleaching and may introduce motion blur in live samples. Higher excitation intensity collects signal faster but accelerates phototoxicity and photobleaching.
The goal is finding the sweet spot where you maximize photon collection within biological constraints. Using the full dynamic range of your camera—avoiding both underexposure and saturation—ensures optimal information capture. Bit depth matters: 16-bit acquisition preserves subtle intensity variations that 8-bit imaging discards.
Sample Preparation Excellence
Many image quality problems originate in sample preparation. Dust and debris on coverslips create dark shadows and bright scatter spots. Bubbles in mounting media cause dramatic optical distortions. Improperly fixed or permeabilized samples produce weak, uneven staining.
Developing rigorous sample preparation protocols with appropriate quality controls prevents systematic artifacts. Simple practices like filtering solutions, using high-quality coverslips, and thoroughly cleaning optical surfaces eliminate common problems. For fluorescence imaging, minimizing background fluorescence through careful reagent selection and thorough washing steps dramatically improves signal-to-noise ratios.
Environmental Stability
Microscopes are sensitive instruments affected by temperature fluctuations, vibration, and air currents. Temperature changes cause focus drift as specimen chambers and microscope components expand or contract. Vibration from nearby equipment or building infrastructure introduces blur and registration errors in time-lapse imaging.
Locating microscopes on isolation tables in temperature-controlled environments represents the gold standard, but practical compromises often suffice. Allowing adequate thermal equilibration time before imaging, using objective heaters for live-cell work, and scheduling critical acquisitions during low-activity building periods improve stability.
🛠️ Post-Acquisition Processing Techniques
Even with optimal acquisition, computational processing plays an essential role in extracting clean signals from noisy measurements.
Flat-Field Correction for Illumination Uniformity
Flat-field correction mathematically compensates for uneven illumination by dividing each image by a reference image of uniform fluorescence. Acquiring proper flat-field references requires careful attention—imaging concentrated fluorescent dye solutions or specialized fluorescent slides while intentionally defocusing to blur out any structure.
The corrected image is calculated as: (Raw Image – Dark Image) / (Flat Field – Dark Image) × Mean Flat Field Value. This simple formula dramatically improves quantitative accuracy and enables reliable intensity comparisons across the entire field of view.
Noise Reduction Filtering
Digital filters can reduce noise while preserving important image features. Gaussian filtering provides straightforward smoothing by averaging pixels with their neighbors, but blurs edges. Median filtering excels at removing salt-and-pepper noise while preserving edges. More sophisticated approaches like bilateral filtering and non-local means denoising achieve superior noise reduction with minimal feature loss.
The key principle is matching filter characteristics to noise properties. Aggressive filtering risks removing real biological structures or creating artificial smoothness that compromises downstream analysis. Applying filters conservatively and validating results against unfiltered data helps avoid over-processing artifacts.
Deconvolution for Optical Sectioning
Deconvolution algorithms use knowledge of the microscope’s point spread function to computationally remove out-of-focus blur and reassign light to its correct spatial origin. This technique can transform widefield images, dramatically improving contrast and resolution.
Multiple deconvolution algorithms exist, ranging from simple nearest-neighbor deblurring to computationally intensive maximum likelihood and Bayesian approaches. Iterative methods like Richardson-Lucy deconvolution provide excellent results but require accurate point spread functions and appropriate stopping criteria to avoid over-processing artifacts.
🤖 Machine Learning Approaches to Image Restoration
Recent advances in deep learning have revolutionized image denoising and artifact correction. Neural networks trained on pairs of noisy and clean images learn to predict the underlying clean signal from corrupted observations.
Content-Aware Image Restoration Networks
Convolutional neural networks like CARE (Content-Aware Image Restoration) and Noise2Noise can achieve remarkable denoising performance that surpasses traditional filtering approaches. These methods learn the statistical properties of both signal and noise, enabling intelligent separation that preserves fine structures while aggressively reducing noise.
The requirement for training data represents both a challenge and an opportunity. Acquiring matched noisy-clean image pairs for training may seem impractical, but creative approaches like imaging the same field at high and low signal-to-noise ratios, or using synthetic noise addition to clean images, provide viable training datasets.
Virtual Staining and Modality Transformation
Taking deep learning further, some networks can predict fluorescence images from label-free modalities like brightfield or phase contrast. This virtual staining approach potentially reduces phototoxicity and photobleaching by minimizing fluorescence exposure while maintaining information content.
These techniques remain active research areas with important validation requirements. Ensuring that network predictions accurately represent biological reality rather than plausible-but-incorrect hallucinations requires rigorous controls and biological validation.
📊 Quality Control and Dataset Validation
Maintaining dataset quality requires systematic quality control measures throughout the imaging pipeline.
Quantitative Image Quality Metrics
Objective metrics help assess image quality and detect problematic images within large datasets. Signal-to-noise ratio quantifies the relative strength of signal versus noise. Contrast-to-noise ratio measures the detectability of features against background. Focus quality metrics detect out-of-focus images that should be excluded from analysis.
Implementing automated quality control that flags images falling below threshold metrics prevents low-quality data from contaminating downstream analysis. Recording quality metrics for each image enables retrospective troubleshooting and systematic identification of factors affecting image quality.
Validation Against Ground Truth
When applying noise reduction or artifact correction, validating results against known ground truth ensures that processing improves rather than distorts data. Using synthetic images with known properties, phantom samples with characterized structures, or comparing processed images against higher-quality reference modalities provides confidence in correction accuracy.
Resolution targets, fluorescent bead slides, and standardized reference samples enable objective quality assessment and facilitate comparison across time, instruments, and laboratories. Incorporating these controls into regular imaging workflows supports quality assurance and helps detect equipment issues before they compromise valuable experiments.
💡 Practical Workflow Integration
Implementing these techniques effectively requires integrating them into coherent workflows adapted to specific research needs.
Start by characterizing your imaging system’s noise properties and systematic artifacts under your typical operating conditions. Acquire reference images including dark frames, flat fields, and point spread functions that enable correction algorithms. Establish standard operating procedures for sample preparation, microscope operation, and image acquisition that minimize variability.
Choose processing approaches appropriate to your noise characteristics and biological questions. Simple filtering may suffice for high-quality images, while demanding applications like single-molecule tracking or weak-signal quantification may require advanced denoising or deconvolution. Document all processing steps and parameters to ensure reproducibility and enable methods sections in publications.
Consider computational resources and throughput requirements. Some advanced techniques require substantial processing time that may be impractical for high-throughput screening with thousands of images. Cloud computing and GPU acceleration can alleviate bottlenecks, but introduce cost and data management considerations.
🔍 Future Perspectives: Emerging Technologies
The field continues evolving rapidly. Computational microscopy approaches increasingly blur the distinction between optical and digital processing, with acquisition and computation jointly optimized. Adaptive imaging systems that adjust parameters in real-time based on sample properties promise more efficient data collection.
New camera technologies with improved sensitivity and reduced noise continue emerging. Advanced neural network architectures achieve better denoising with less training data. Physics-informed machine learning that incorporates optical principles into network architectures may overcome current limitations of pure data-driven approaches.
Open-source tools and standardized file formats facilitate sharing and comparison of methods across the research community. Initiatives promoting imaging data standards and computational reproducibility help ensure that advances in image quality translate to more reliable scientific conclusions.
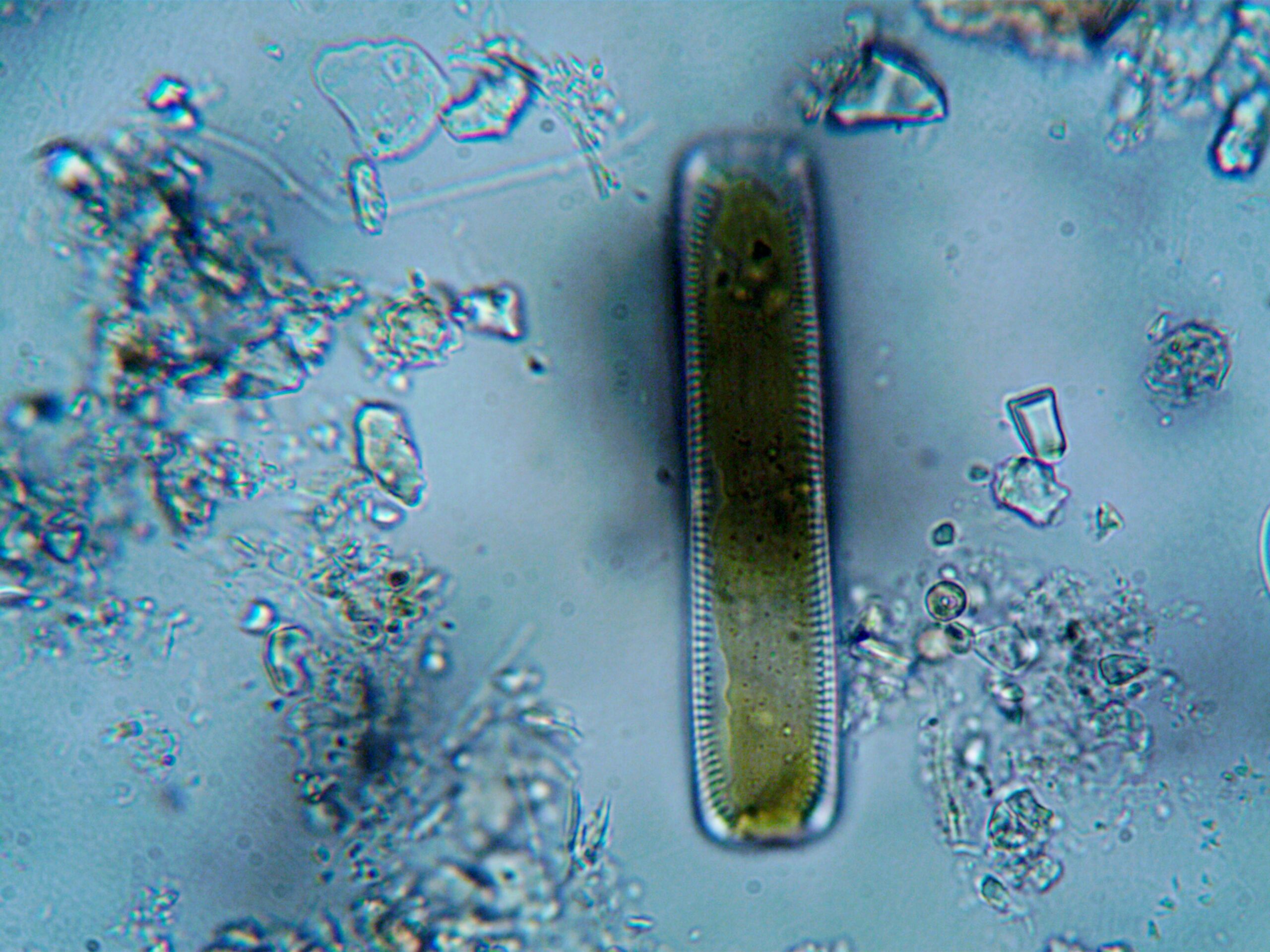
🎓 Building Expertise and Community Resources
Mastering cellular image quality requires ongoing learning and engagement with the microscopy community. Online forums, workshops, and courses provide opportunities to develop skills and learn from others’ experiences. Many research institutions maintain imaging core facilities with expert staff who can provide guidance on optimizing image acquisition and processing for specific applications.
Open-source software packages like ImageJ/Fiji, CellProfiler, and napari provide accessible platforms for implementing quality control and correction workflows. Python and MATLAB ecosystems offer extensive image processing libraries and active communities contributing new methods. Investing time in developing computational skills pays dividends in research productivity and data quality.
Documenting your own protocols and sharing them with colleagues builds institutional knowledge and improves reproducibility. Creating image quality benchmarks specific to your research questions enables systematic evaluation of new methods and instruments. Treating image quality as a first-class research consideration rather than a technical afterthought elevates the reliability of biological conclusions.
Crystal-clear cellular images don’t happen by accident. They result from deliberate attention to acquisition parameters, systematic artifact correction, and thoughtful application of computational tools. By understanding noise sources, recognizing common artifacts, and implementing appropriate mitigation strategies, researchers transform messy datasets into reliable foundations for biological discovery. The investment in image quality expertise directly translates to more accurate measurements, more confident conclusions, and ultimately more impactful science.
Toni Santos is a biological systems researcher and forensic science communicator focused on structural analysis, molecular interpretation, and botanical evidence studies. His work investigates how plant materials, cellular formations, genetic variation, and toxin profiles contribute to scientific understanding across ecological and forensic contexts. With a multidisciplinary background in biological pattern recognition and conceptual forensic modeling, Toni translates complex mechanisms into accessible explanations that empower learners, researchers, and curious readers. His interests bridge structural biology, ecological observation, and molecular interpretation. As the creator of zantrixos.com, Toni explores: Botanical Forensic Science — the role of plant materials in scientific interpretation Cellular Structure Matching — the conceptual frameworks behind cellular comparison and classification DNA-Based Identification — an accessible view of molecular markers and structural variation Toxin Profiling Methods — understanding toxin behavior and classification through conceptual models Toni's work highlights the elegance and complexity of biological structures and invites readers to engage with science through curiosity, respect, and analytical thinking. Whether you're a student, researcher, or enthusiast, he encourages you to explore the details that shape biological evidence and inform scientific discovery.




