Cell shape variability represents one of the most intriguing puzzles in modern biological research, challenging scientists to decode how and why cells adopt different morphologies across tissues and conditions.
🔬 The Fundamental Nature of Cellular Morphology
Cells are the building blocks of life, yet they display remarkable diversity in their physical appearance. From the elongated neurons stretching across our nervous system to the perfectly rounded red blood cells coursing through our veins, cellular architecture varies dramatically. This morphological diversity isn’t merely aesthetic—it’s fundamentally linked to cellular function, survival, and the overall health of organisms.
Understanding cell shape variability has become increasingly critical as researchers recognize that changes in cellular morphology often precede or accompany disease states. Cancer cells, for instance, frequently exhibit abnormal shapes that distinguish them from healthy tissue. Neurological disorders can manifest through altered neuronal morphology, while cardiovascular diseases often involve changes in endothelial cell shape.
The challenge lies not just in observing these variations but in accurately matching cellular phenotypes across different experimental conditions, time points, and research methodologies. This matching problem has emerged as a significant bottleneck in cellular research, limiting our ability to draw meaningful conclusions from vast amounts of microscopy data.
Why Cell Shape Matters More Than You Think 🧬
Cellular morphology serves as a visual readout of internal cellular processes. The cytoskeleton—a dynamic network of protein filaments—actively shapes cells in response to biochemical signals, mechanical forces, and environmental conditions. When these signals change, so does the cell shape, making morphology a sensitive indicator of cellular state.
In tissue engineering and regenerative medicine, controlling cell shape has proven essential for directing stem cell differentiation. Researchers have discovered that simply changing the geometric constraints on stem cells can push them toward becoming bone cells, fat cells, or other specialized types. This mechanobiological principle underscores how form and function are inseparably linked at the cellular level.
Drug development also relies heavily on analyzing cell shape changes. Pharmaceutical companies screen thousands of compounds by examining how they affect cellular morphology. A drug that causes abnormal cell shapes might indicate toxicity, while compounds that restore normal morphology in disease models could represent promising therapeutic candidates.
The Matching Challenge: Where Complexity Meets Technology
The central challenge in studying cell shape variability lies in matching—comparing cells across different conditions, identifying similar morphological phenotypes, and tracking cellular changes over time. This seemingly straightforward task becomes extraordinarily complex when dealing with the natural heterogeneity of cell populations.
Even genetically identical cells cultured under identical conditions display considerable morphological variation. This intrinsic variability makes it difficult to determine whether observed differences between experimental groups represent true biological effects or simply reflect the natural range of cellular shapes.
Quantifying the Unquantifiable
Traditional approaches to cell shape analysis relied on simple metrics like circularity, aspect ratio, or area. While useful for basic categorization, these parameters fail to capture the rich complexity of cellular morphology. A cell might have the same circularity value as another yet possess completely different boundary characteristics, texture patterns, or internal organization.
Modern computational approaches have introduced sophisticated shape descriptors including:
- Fourier descriptors that decompose cell boundaries into frequency components
- Zernike moments that capture rotational invariant shape features
- Morphological profiles that measure multiple cellular characteristics simultaneously
- Graph-based representations that encode spatial relationships between cellular components
- Deep learning features extracted from convolutional neural networks trained on cellular images
Each method offers unique advantages but also introduces new challenges in interpretation, reproducibility, and biological relevance.
⚡ High-Throughput Microscopy: Data Abundance and Analysis Scarcity
Advances in automated microscopy have revolutionized our ability to image cells. Modern high-content screening platforms can capture millions of cellular images in single experiments, generating datasets that would have been unimaginable just a decade ago. This technological leap has shifted the bottleneck from data acquisition to data analysis.
The matching problem intensifies with scale. When comparing tens of thousands of cells across multiple experimental conditions, researchers need robust statistical frameworks that account for both biological variability and technical noise. Standard parametric statistical tests often fail because cellular morphology data rarely follows normal distributions and frequently violates assumptions of independence.
Machine Learning Enters the Arena
Artificial intelligence and machine learning have emerged as powerful tools for addressing cell shape matching challenges. Supervised learning algorithms can be trained to classify cells into predefined categories based on morphological features. Once trained, these models can process new images rapidly, providing consistent classifications that would be impossible through manual analysis.
Unsupervised learning approaches offer even more intriguing possibilities. Clustering algorithms can discover natural groupings within cell populations without requiring predefined categories. This exploratory approach has revealed previously unrecognized cellular subtypes and transitional states that challenge traditional classification schemes.
Deep learning, particularly convolutional neural networks, has achieved remarkable success in cellular image analysis. These networks can learn hierarchical representations of cell shape directly from raw images, potentially capturing morphological patterns too subtle or complex for human observers to articulate.
Biological Variability Versus Technical Artifacts 🔍
One of the most persistent challenges in matching cell shapes across experiments involves distinguishing genuine biological variability from technical artifacts. Factors like imaging conditions, sample preparation methods, cell culture protocols, and even the time of day when experiments are performed can introduce systematic biases that confound morphological analysis.
Batch effects—systematic differences between experiments performed at different times or locations—pose particular problems. A cell population imaged with one microscope configuration might appear systematically different from the same population imaged with different settings, even though no biological difference exists.
Standardization Efforts and Reproducibility
The research community has increasingly recognized the need for standardized protocols in cellular imaging and analysis. Initiatives like the QUAREP-LiMi (Quality Assessment and Reproducibility for Instruments and Images in Light Microscopy) consortium work to establish best practices for image acquisition, quality control, and data reporting.
Reference datasets with well-characterized cell populations serve as benchmarks for validating new analysis methods. By testing algorithms against these standardized datasets, researchers can more objectively evaluate performance and identify methods that generalize well across different experimental contexts.
Temporal Dynamics: Matching Cells Across Time ⏱️
Cell shape isn’t static—it changes continuously as cells respond to stimuli, progress through the cell cycle, migrate, or differentiate. Time-lapse microscopy captures these dynamic processes, but analyzing temporal sequences introduces additional matching challenges.
Tracking individual cells across time points requires solving correspondence problems: which cell in frame t+1 is the same as a particular cell in frame t? This becomes especially difficult in crowded fields where cells overlap, divide, or migrate out of the imaging area.
Advanced tracking algorithms employ probabilistic models that consider both morphological similarity and spatial proximity. Kalman filters, particle filters, and graph-based optimization methods attempt to maintain cell identities across frames while accounting for uncertainty and occasional tracking errors.
Multi-Dimensional Phenotyping: Beyond Simple Shape 🎯
Modern cellular research increasingly recognizes that shape represents just one dimension of cellular phenotype. Comprehensive characterization requires integrating morphological data with fluorescent markers indicating protein localization, gene expression patterns, metabolic activity, and functional responses.
This multi-dimensional phenotyping creates even more complex matching problems. Researchers must now align cells not just based on shape but on combinations of dozens or hundreds of features spanning different measurement modalities. High-dimensional data analysis techniques like t-SNE, UMAP, and manifold learning help visualize and interpret these complex phenotypic spaces.
Single-Cell Resolution Meets Population-Level Analysis
The tension between single-cell precision and population-level statistics represents another facet of the matching challenge. Individual cells might display unusual morphologies that could represent either rare but biologically important states or irrelevant outliers. Distinguishing signal from noise at the single-cell level requires sophisticated statistical frameworks that can handle extreme data heterogeneity.
Meta-analysis approaches that combine data across multiple studies face compounded matching challenges. Different research groups use different cell lines, imaging protocols, and analysis pipelines, making direct comparison difficult. Developing harmonization methods that enable meaningful cross-study comparisons remains an active area of methodological research.
🚀 Emerging Solutions and Future Directions
Despite the formidable challenges, the field is advancing rapidly. Several promising approaches are transforming how researchers address cell shape matching problems.
Transfer Learning and Foundation Models
Transfer learning allows models trained on large general datasets to be fine-tuned for specific cellular analysis tasks with limited labeled data. Foundation models—very large neural networks trained on diverse cellular images—can serve as universal feature extractors, providing robust shape representations that generalize across different cell types and imaging conditions.
These models essentially learn a common “language” for describing cellular morphology that remains meaningful across different experimental contexts, potentially solving many matching challenges that arise from technical variability.
Generative Models and Synthetic Data
Generative adversarial networks (GANs) and variational autoencoders (VAEs) can learn the distribution of cell shapes and generate synthetic cellular images. These synthetic datasets serve multiple purposes: augmenting training data for machine learning models, testing analysis pipelines under controlled conditions, and potentially standardizing cellular phenotypes by transforming images to remove batch effects while preserving biological information.
Practical Implications for Research and Medicine 💊
Solving the cell shape matching challenge has immediate practical implications. In clinical diagnostics, improved morphological analysis could enable earlier disease detection and more accurate prognosis. Pathologists already use cellular morphology to diagnose cancer and other conditions, but computational methods could enhance sensitivity and objectivity.
Personalized medicine could benefit from matching patient-derived cells against extensive databases of morphological phenotypes associated with different disease states and treatment responses. This morphological profiling might help predict which patients will respond to specific therapies or identify individuals at high risk for adverse effects.
In drug discovery, better matching methods would accelerate compound screening by more reliably identifying morphological changes associated with desired therapeutic effects. This could reduce false positives that waste resources in clinical development and false negatives that cause potentially valuable drugs to be discarded.
Collaborative Science and Open Resources 🌐
Addressing the matching challenge requires collaboration across disciplines—biologists, computer scientists, statisticians, and clinicians must work together to develop solutions that are both technically sophisticated and biologically meaningful. Open science practices, including sharing datasets, code, and protocols, accelerate progress by allowing researchers worldwide to build on each other’s work.
Public repositories like the Image Data Resource (IDR) and Cell Image Library provide access to vast collections of cellular images. Challenge competitions, where researchers compete to solve specific analysis problems using common datasets, have proven effective for driving methodological innovation while providing objective performance benchmarks.
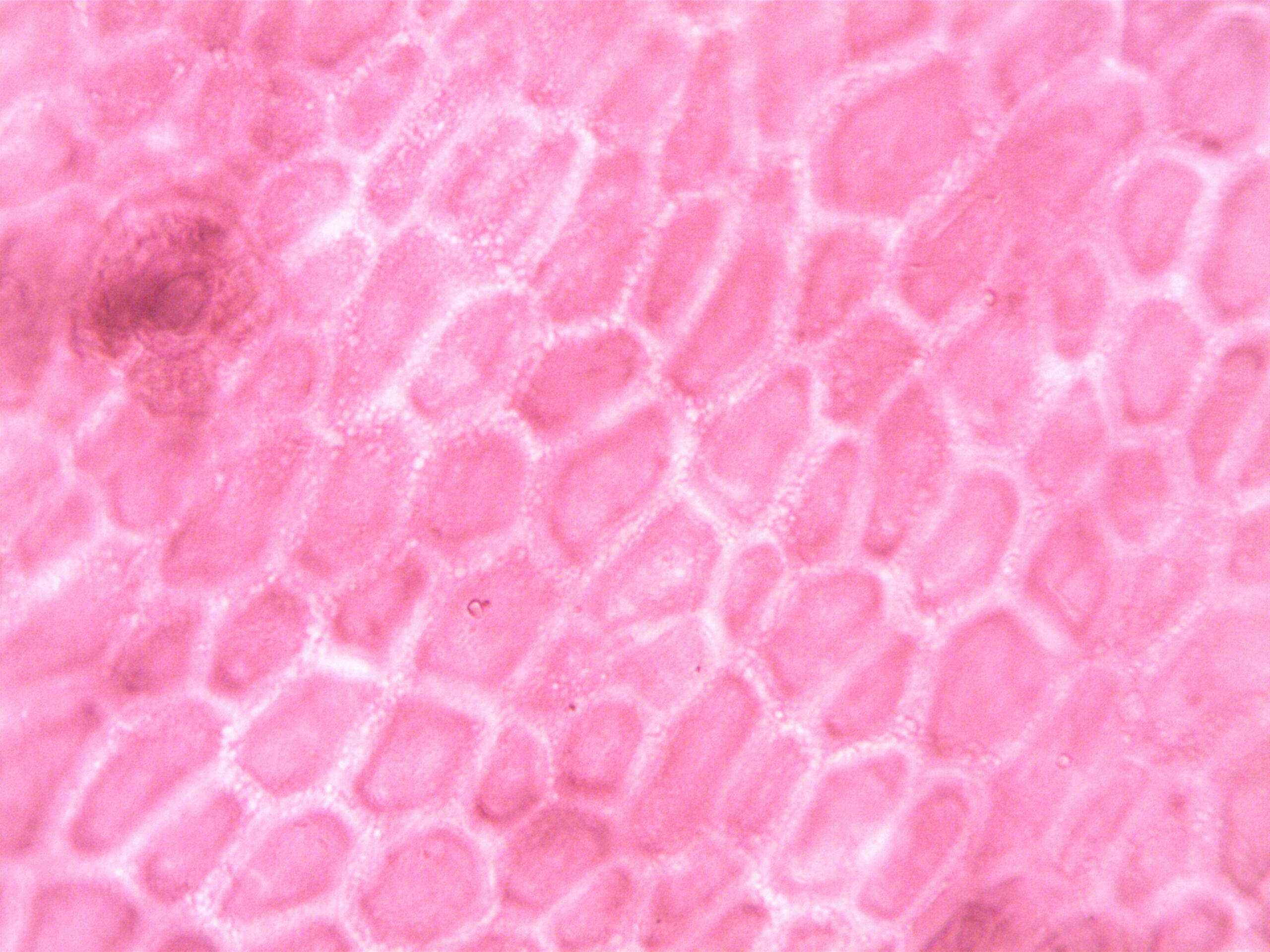
Navigating Complexity Toward Understanding 🎓
The challenge of matching cell shapes across different conditions reflects broader themes in modern biology: managing complexity, extracting meaning from massive datasets, and integrating diverse sources of information. As technologies continue to advance, the ability to precisely quantify and compare cellular morphology will increasingly become a cornerstone of biological research.
Success in this domain requires balancing sophistication with interpretability. The most powerful computational methods mean little if researchers cannot understand what morphological features drive classifications or predictions. Explainable AI approaches that provide insights into model decisions are essential for maintaining biological interpretability alongside computational power.
The journey from observing cell shape variability to truly understanding its biological significance continues. Each methodological advance reveals new layers of complexity while simultaneously providing tools to address those complexities. The matching challenge, far from being merely technical, lies at the heart of decoding cellular function and dysfunction.
As we develop increasingly sophisticated tools for analyzing cell shape, we move closer to answering fundamental questions about how cells sense their environment, coordinate their activities with neighbors, respond to perturbations, and ultimately give rise to the extraordinary complexity of living organisms. The mystery of cell shape variability gradually yields to persistent inquiry, technological innovation, and collaborative scientific effort.
Toni Santos is a biological systems researcher and forensic science communicator focused on structural analysis, molecular interpretation, and botanical evidence studies. His work investigates how plant materials, cellular formations, genetic variation, and toxin profiles contribute to scientific understanding across ecological and forensic contexts. With a multidisciplinary background in biological pattern recognition and conceptual forensic modeling, Toni translates complex mechanisms into accessible explanations that empower learners, researchers, and curious readers. His interests bridge structural biology, ecological observation, and molecular interpretation. As the creator of zantrixos.com, Toni explores: Botanical Forensic Science — the role of plant materials in scientific interpretation Cellular Structure Matching — the conceptual frameworks behind cellular comparison and classification DNA-Based Identification — an accessible view of molecular markers and structural variation Toxin Profiling Methods — understanding toxin behavior and classification through conceptual models Toni's work highlights the elegance and complexity of biological structures and invites readers to engage with science through curiosity, respect, and analytical thinking. Whether you're a student, researcher, or enthusiast, he encourages you to explore the details that shape biological evidence and inform scientific discovery.




